Biological systems are exceedingly complex: they are multi-scale, nonlinear, and subject to random variability at the molecular level. This complexity means that computer models are vital for uncovering mechanistic principles based on various types of experimental data. Current projects relate to:
Biomolecular Network Dynamics
 Underlying all cellular processes are complex networks of biochemical reactions. An ever-expanding wealth of data presents new opportunities to infer these networks, understand their dynamics, and gain quantitative understanding of cellular function. For example, quantitative models of gene regulatory network dynamics are helping to reveal how cell identity—or phenotype—is achieved, maintained, and reprogrammed, with applications in stem cell engineering and regenerative medicine.
Underlying all cellular processes are complex networks of biochemical reactions. An ever-expanding wealth of data presents new opportunities to infer these networks, understand their dynamics, and gain quantitative understanding of cellular function. For example, quantitative models of gene regulatory network dynamics are helping to reveal how cell identity—or phenotype—is achieved, maintained, and reprogrammed, with applications in stem cell engineering and regenerative medicine.
We are advancing techniques for modeling complex gene regulatory networks and simulating their dynamic behavior. One challenge in this area is the curse-of-dimensionality, wherein the computational cost of modeling complex network dynamics grows exponentially with the number of molecular species. To address this challenge, we are developing modeling techniques that balance computational expense with accuracy in treatment of the single-molecule-level reaction kinetics.
Stochastic Modeling and Simulation
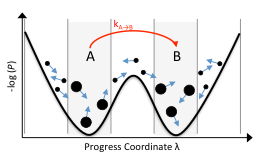 Experimental techniques are increasingly incisive, revealing information at single-cell, single-nucleotide, or single-molecule resolution. These experiments have revealed often-unexpected levels of heterogeneity at these scales. In order to quantitatively understand this heterogeneity, stochastic models are required. We advance probabilistic modeling techniques, with a theoretical underpinning of stochastic chemical kinetics.
Experimental techniques are increasingly incisive, revealing information at single-cell, single-nucleotide, or single-molecule resolution. These experiments have revealed often-unexpected levels of heterogeneity at these scales. In order to quantitatively understand this heterogeneity, stochastic models are required. We advance probabilistic modeling techniques, with a theoretical underpinning of stochastic chemical kinetics.
We are developing simulation techniques that can reduce the computational expense of simulating complex systems. Specifically, we develop rare-event sampling techniques and apply them in a number of areas, from dynamics of cell differentiation and reprogramming, to spatial reorganization of proteins on cell surfaces.
Cell Fate-Decisions
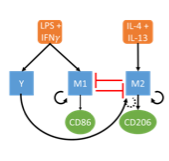 An overarching question in biology is how cells integrate a variety of environmental signals and execute a particular phenotypic or functional response. For example, in the development of multicellular organisms, cells with the same DNA must differentiate into tissue-specific cell-types. As another example, cells of the immune system coordinate levels of inflammation in response to a variety of cues. These processes are highly complex, linking extra-cellular events, intracellular signal transduction, transcriptional networks, and epigenetic regulation.
An overarching question in biology is how cells integrate a variety of environmental signals and execute a particular phenotypic or functional response. For example, in the development of multicellular organisms, cells with the same DNA must differentiate into tissue-specific cell-types. As another example, cells of the immune system coordinate levels of inflammation in response to a variety of cues. These processes are highly complex, linking extra-cellular events, intracellular signal transduction, transcriptional networks, and epigenetic regulation.
We develop mathematical models in collaboration with experimentalists to tackle this complexity and advance understanding of cell fate-decisions. These projects draw heavily on parameter inference and model selection techniques. Current areas of focus include macrophage polarization and DNA methylation maintenance.
